Catalogue
SCICONS anti-dsRNA antibody (J2)
Catalog number: 10010200$541.00
Add To Cart| Clone | J2 |
| Isotype | IgG2a kappa |
| Product Type |
Monoclonal Antibody |
| Units | 200 µg |
| Host | Mouse |
| Application |
Dot blot dsRNA-immunoblotting ELISA Flow Cytometry Immuno-affinity-chromatography Immunocytochemistry Immunohistochemistry |
Background
This product is part of the SCICONS™ product line which offers the gold standard in anti-dsRNA antibodies.
Over the past decade our double-stranded RNA (dsRNA)antibodies have been used extensively to detect and characterise plant and animal viruses with dsRNA genomes or intermediates. In addition, the anti-dsRNA antibodies can be used as a diagnostic tool to detect pathogens, including detection in paraffin-embedded fixed tissue samples (Richardson et al. 2010).
The J2 anti-dsRNA IgG2a monoclonal antibody has become the gold standard in dsRNA detection. It was used initially for the study of plant viruses, but since the seminal paper of Weber et al. in 2006, where J2 was used to show that all the positive strand RNA viruses tested produced copious amounts of dsRNA in infected cells, this antibody has been used extensively in a wide range of systems, as documented in over 200 scientific publications.
J2 can be used to detect dsRNA intermediates of viruses as diverse as Hepatitis C virus, Dengue virus, rhinovirus, Chikungunya virus, Rabies virus, Polio virus, Classic swine fever virus, Brome mosaic virus and many more in cultured cells and also in fixed paraffin-embedded histological samples.
J2 has been used to elucidate how anti-viral responses are initiated, what counter-strategies viruses have adopted to avoid them, and to explore the viral life cylce by enabling ultrastructiural localisation studies of viral nucleic acid replication sites (Welsch et al., 2009 & Knoops et al., 2011).
J2 has been used successfully in electron microscopy, in immunofluorescence microscopy, in immunohistochemistry, and various immunocapture methods, such as dot blots and ELISA. J2 has also been recommended as a diagnostic tool to detect whether an unkown pathogen is bacterial or viral in nature (Richardson et al., 2010). Recently J2 has also been used to monitor the removal of dsRNA from in vitro synthethised mRNA preparations that may have potential use in gene therapy (Kariko et al., 2011).
Synonyms: Mouse anti dsRNA
Source
Female DBA/2 mice were injected intraperitonially with a mixture of 50 ug L-dsRNA and 75 ug methylated bovine serum albumin, emulsified in complete Freund's adjuvant. After several boosts spleen cells were fused with Sp2/0-Agl4 myeloma cells to generate the hybridoma clone.
Product
Mouse monoclonal antibody J2 recognises double-stranded RNA (dsRNA) provided that the length of the helix is greater than or equal to 40 bp. dsRNA-recognition is independent of the sequence and nucleotide composition of the antigen. All naturally occurring dsRNAs investigated up to now (40-50 species) as well as poly(I).poly(C) and poly(A).poly(U) have been recognised by J2, although in some assays its affinity to poly(I).poly(C) is about 10 times lower than that to other dsRNA antigens
Formulation: The lyophilised sample should be reconstituted with 200 µl sterile distilled water. The mAb will then be in PBS without any stabilisers or preservatives at a concentration of 1 mgr/ml. As a result of the lyophilisation procedure, the reconstituted antibody may contain small amounts of denatured protein in the form of aggregates that may interfere with some applications such as immunohistochemistry (e.g. by giving high backgrounds). We therefore highly recommend centrifuging (microcentrifuge) the reconstituted antibody before use and using the supernatant.
Purification Method: Affinity chromatography on Protein G.
Purity: Gel electrophoretically pure IgG antibody.
Concentration: Concentration after reconstitution: 1.00 mg/ml as determined by A280 nm (A280 nm = 1.47 corresponds to 1 mg/ml antibody).
Applications
Mouse monoclonal antibody J2 can be used for ELISA, dsRNA-immunoblotting, immunoaffinity chromatography and in certain systems also for immunohistochemistry (see references). The optimum working dilution of the antibody for any specific application should be established by titration. Also the optimal incubation time and temperature should be established for each specific application.
Please note that nucleic acid separation prior to dsRNA-immunoblotting must be carried out by polyacrylamide gel electrophoresis, because the sensitivity of detection is considerably lower after blotting from agarose gels.
Not for use for clinical purposes. For in vitro use only.
Storage
After reconstitution antibodies should be aliquoted and stored at -20 °C or -70°C. After adding 10 mM sodium azide undiluted antibody can also be stored at +4 °C for a short period of time. For long term storage the mAb should be kept frozen. Repeated freezing/thawing cycles should be avoided. When kept lyophilized the product will remain stable for 10 years at -20 °C or -70°C.
Shipping Conditions: Ship at ambient temperature.
Caution
This product is intended FOR RESEARCH USE ONLY, and FOR TESTS IN VITRO, not for use in diagnostic or therapeutic procedures involving humans or animals. It may contain hazardous ingredients. Please refer to the Safety Data Sheets (SDS) for additional information and proper handling procedures. Dispose product remainders according to local regulations.This datasheet is as accurate as reasonably achievable, but our company accepts no liability for any inaccuracies or omissions in this information.
References
1) F. Weber, V. Wagner, S. B. Rasmussen, R. Hartmann, S. R. Paludan. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol (2006), 80(10):5059-64. doi: 10.1128/JVI.80.10.5059-5064.2006.
2) S. Welsch, S. Miller, I. Romero-Brey, A. Merz, C. K. E. Bleck, P. Walther, S. D. Fuller, C. Antony, J. Krijnse-Locker, R. Bartenschlager. Composition and Three-Dimensional Architecture of the Dengue Virus Replication and Assembly Sites. Cell Host & Microbe (2009) 5(4); 365-375. doi.org/10.1016/j.chom.2009.03.007.
3) K. Knoops , M. Bárcena, R. W. Limpens, A. J. Koster, A. M. Mommaas, E. J. Snijder. Ultrastructural characterization of arterivirus replication structures: reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J Virol. (2012) 86(5); 2474-2487. doi:10.1128/JVI.06677-11.
4) S. J. Richardson, A. Willcox, D. A. Hilton, S. Tauriainen, H. Hyoty, A. J. Bone, A. K. Foulis, N. G. Morgan. Use of antisera directed against dsRNA to detect viral infections in formalin-fixed paraffin-embedded tissue. J Clin Virol. (2010) 49(3); 180-5. doi: 10.1016/j.jcv.2010.07.015.
5) K. Karikó, H. Muramatsu, J. Ludwig, D. Weissman, Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA, Nucleic Acids Research (2011) 39(21); e142, https://doi.org/10.1093/nar/gkr695.
6) Schönborn, J., Oberstrass, J., Breyel, E., Tittgen, J., Schumacher, J. and Lukacs, N. (1991) Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res.19, 2993-3000.
7) Lukacs, N. (1994) Detection of virus infection in plants and differentiation between coexisting viruses by monoclonal antibodies to double-stranded RNA. J. Virol. Methods 47, 255-272.
8) Lukacs, N. (1997) Detection of sense:antisense duplexes by structurespecific anti-RNA antibodies. In: Antisense Technology. A Practical Approach, C. Lichtenstein and W. Nellen (eds), pp. 281-295. IRL Press, Oxford.
Recent publication:
Tirosh Shapira, I. Abrrey Monreal, Sébastien P Dion, Mason Jager, Antoine Désilets, Andrea D Olmstead, Thierry Vandal, David W Buchholz, Brian Imbiakha, Guang Gao, Aaleigha Chin, William D Rees, Theodore Steiner, Ivan Robert Nabi, Eric Marsault, Julie Sahler, Avery August, Gerlinde Van de Walle, Gary R Whittaker, Pierre-Luc Boudreault, Hector C Aguilar, Richard Leduc, François Jean.
A novel highly potent inhibitor of TMPRSS2-like proteases blocks SARS-CoV-2 variants of concern and is broadly protective against infection and mortality in mice. bioRxiv 2021.05.03.442520; doi: https://doi.org/10.1101/2021.05.03.442520
https://biorxiv.org/cgi/content/short/2021.05.03.442520v1
Safety Datasheet(s) for this product:
| SDS antibodies without sodium azide Exalpha_V1.0 |
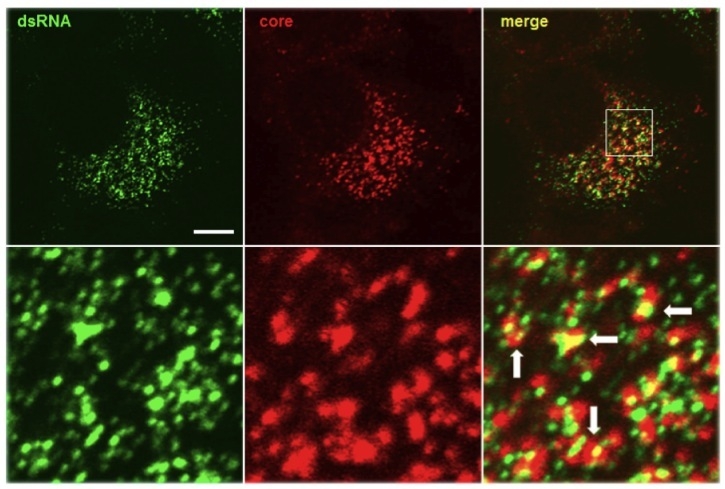
Figure 1. Antibody J2 (1:250 dilution, 4 microgram/ml) reveals the colocalisation of dsRNA (labelled in green) and Hepatitis C virus core protein (labelled in red), indicating the location of putative virus assembly sites (arrows).
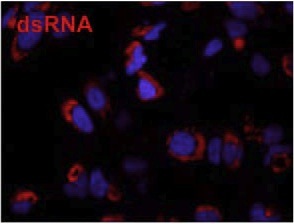
Figure 2. Immunofluorescence microscopy using J2 antibody reveals dsRNA (labelled in red), a marker for the the viral replication complex, in HuH-7 cells infected with Dengue virus. Cellular DNA is labelled with DAPI (blue). Figure taken from Anwar et al. (2011) PLoS One 6:e23246.
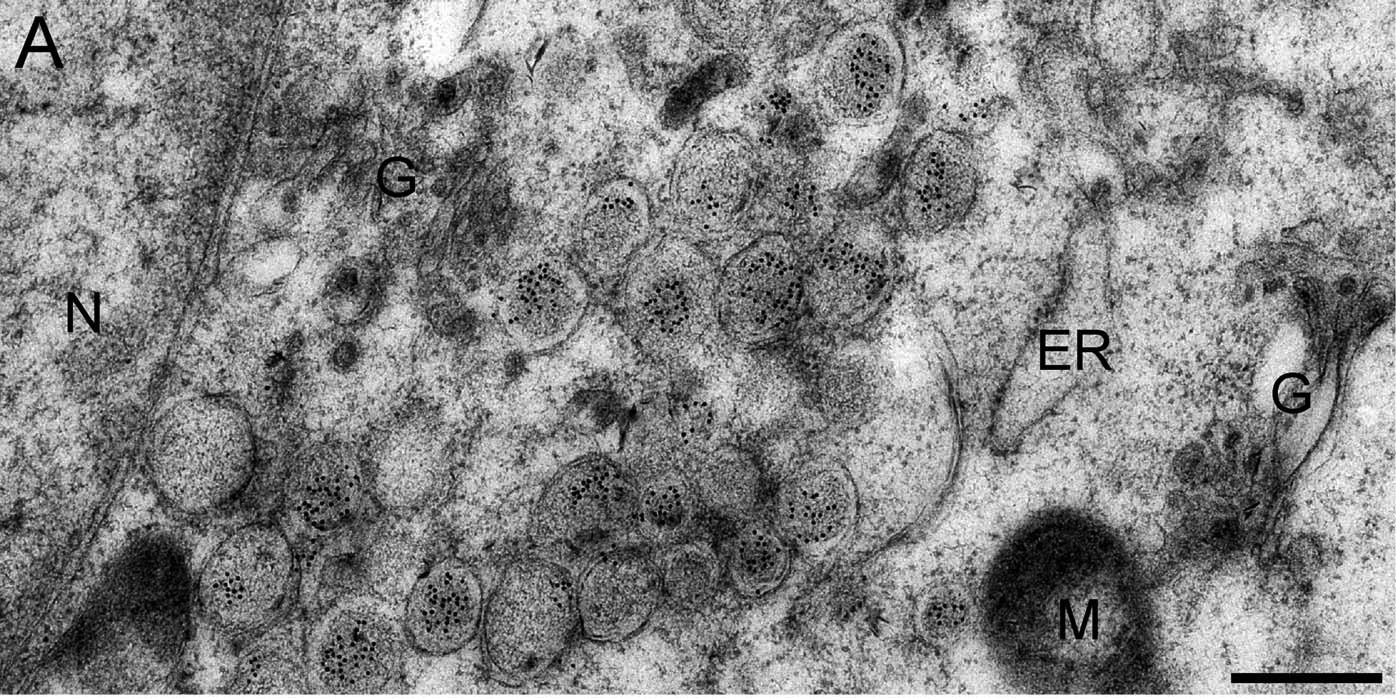
Figure 3. Immune electron microscopy image, where J2 antibody labeling shows abundance of dsRNA (black dots) in double-membrane vesicles (DMVs) of SARS-CoV infected Vero E6 cells, fixed at 7 hours post-infection. J2 was detected using immunogold conjugated to protein A. G: Golgi complex, M: mitochondria, N: nucleus. Figure taken from Knoops et al. (2008) PLoS Biol 6:e226.
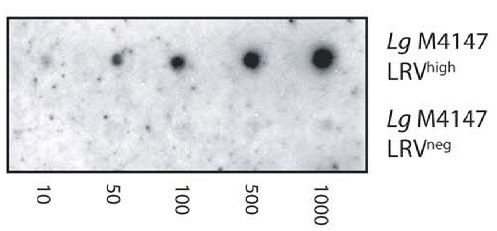
Figure 4. J2 antibody is used in dot blots to detect dsRNA from Leishmania RNA virus (LRV) in the Leishmania parasite L. guyanensis. A serial dilution of 1000 to 10 parasites from LRV-positive and negative control strains - Lg4147LRVhigh and LG4147LRVneg respectively - is shown. Picture taken from Zangger et al. (2013) PLoS Negl Trop Dis 7:e2006.
